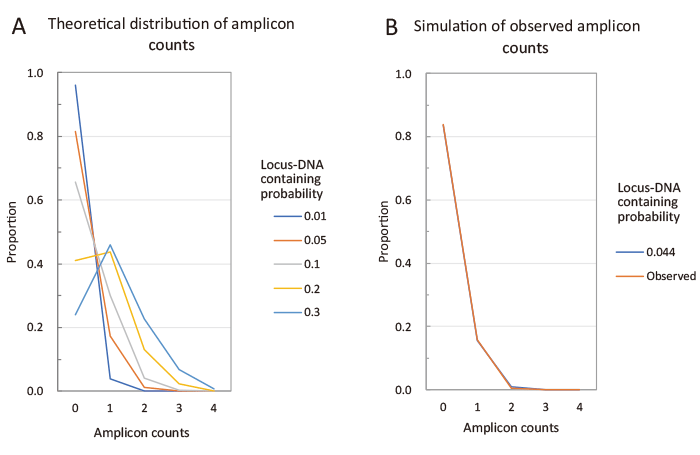
Fig. 3 Simulation of the distribution of the number of amplicons detected in each PCR run. Each PCR run can detect 0–4 different amplicons. The number of amplicons detected per run (amplicon count) depends on the amount of template DNA present in the PCR tube. We calculated the proportion of each amplicon count based on the locus DNA containing probability that 1 μL of whole genome amplification (WGA) eluate used as a PCR template contains one copy of the target locus (denote as C). Detailed calculations are provided in the supplementary materials. Figure A shows the simulated distribution of amplicon counts. As C increases from 0.01 to 0.3, the distribution shifts from a right-skewed shape to one with a peak near 1. Figure B shows the distribution observed in this study. This observed distribution closely matches the simulated distribution with C = 0.044, for which the residual sum of squares is less than 0.0001 (Supplementary materials). To simplify the calculation, the following assumptions were made: (1) Each embryo has four different alleles. (2) PCR amplification from a single copy of DNA always succeeds.